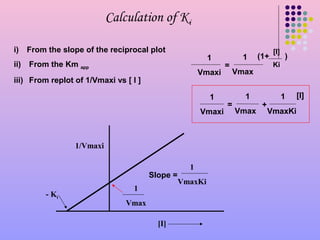
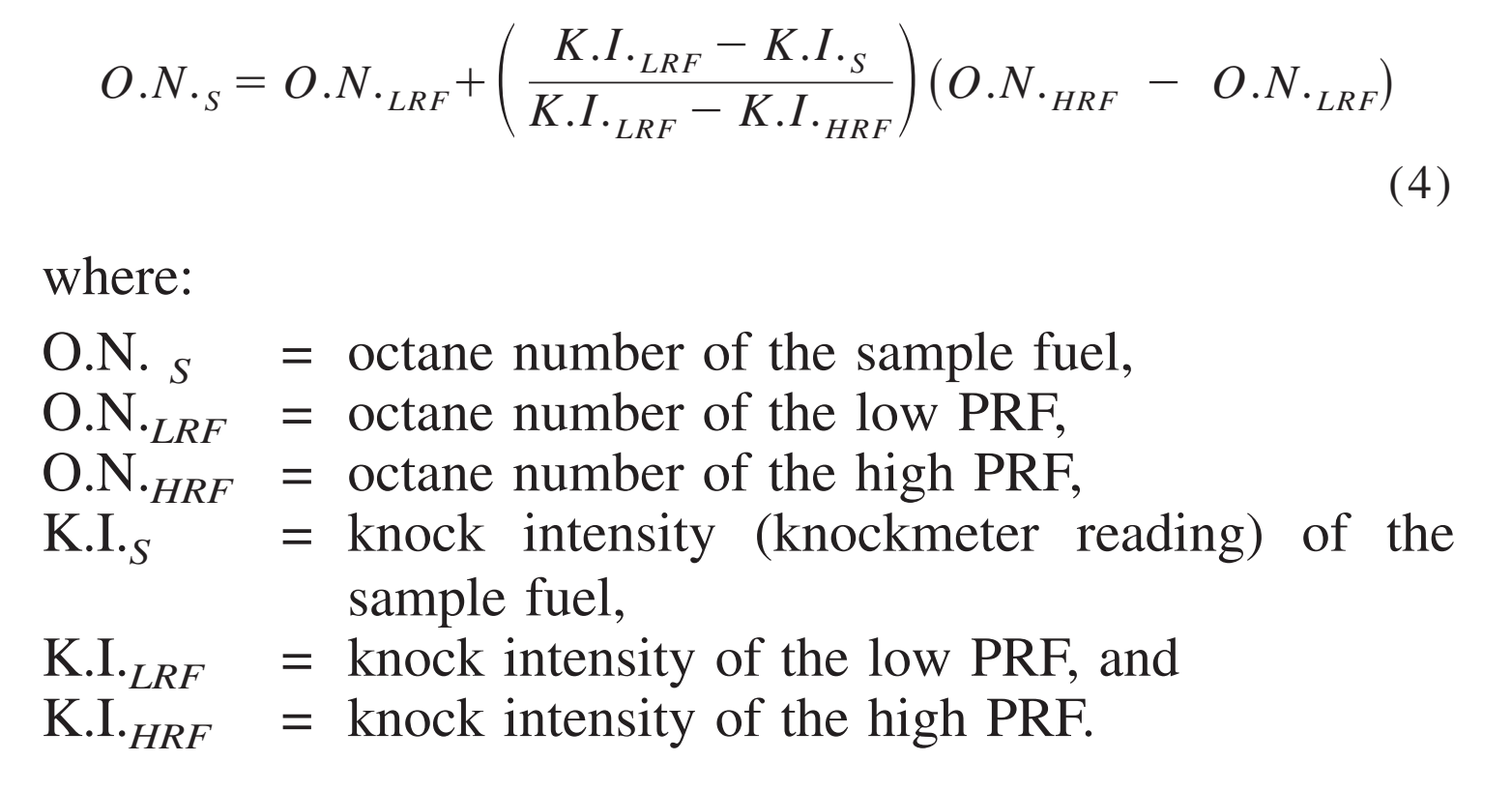
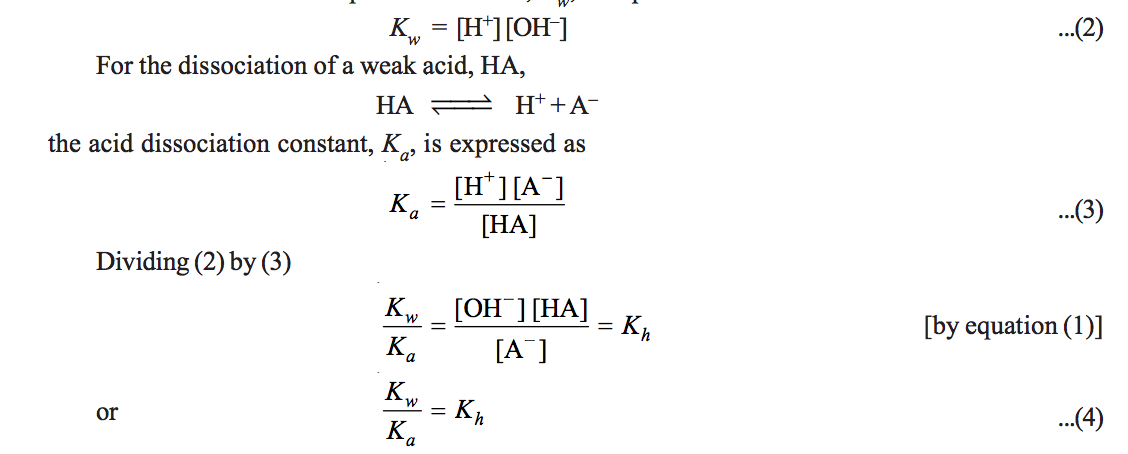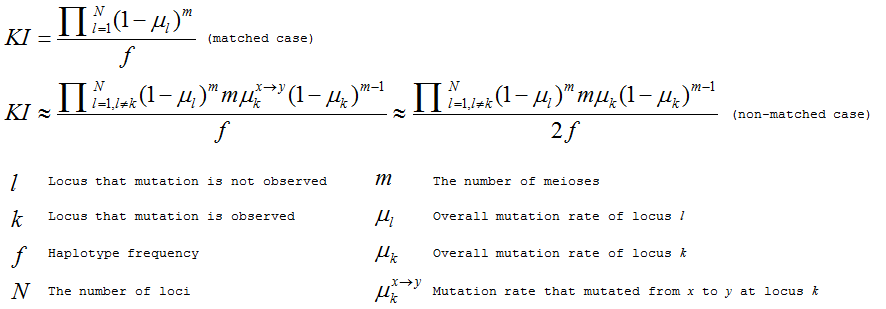Calculate K(c ) for the reaction KI+I(2) hArr KI(3). Given that initial weight of KI is 1.326 g weight of KI(3) is 0.105 g and number of moles of free I(2) is
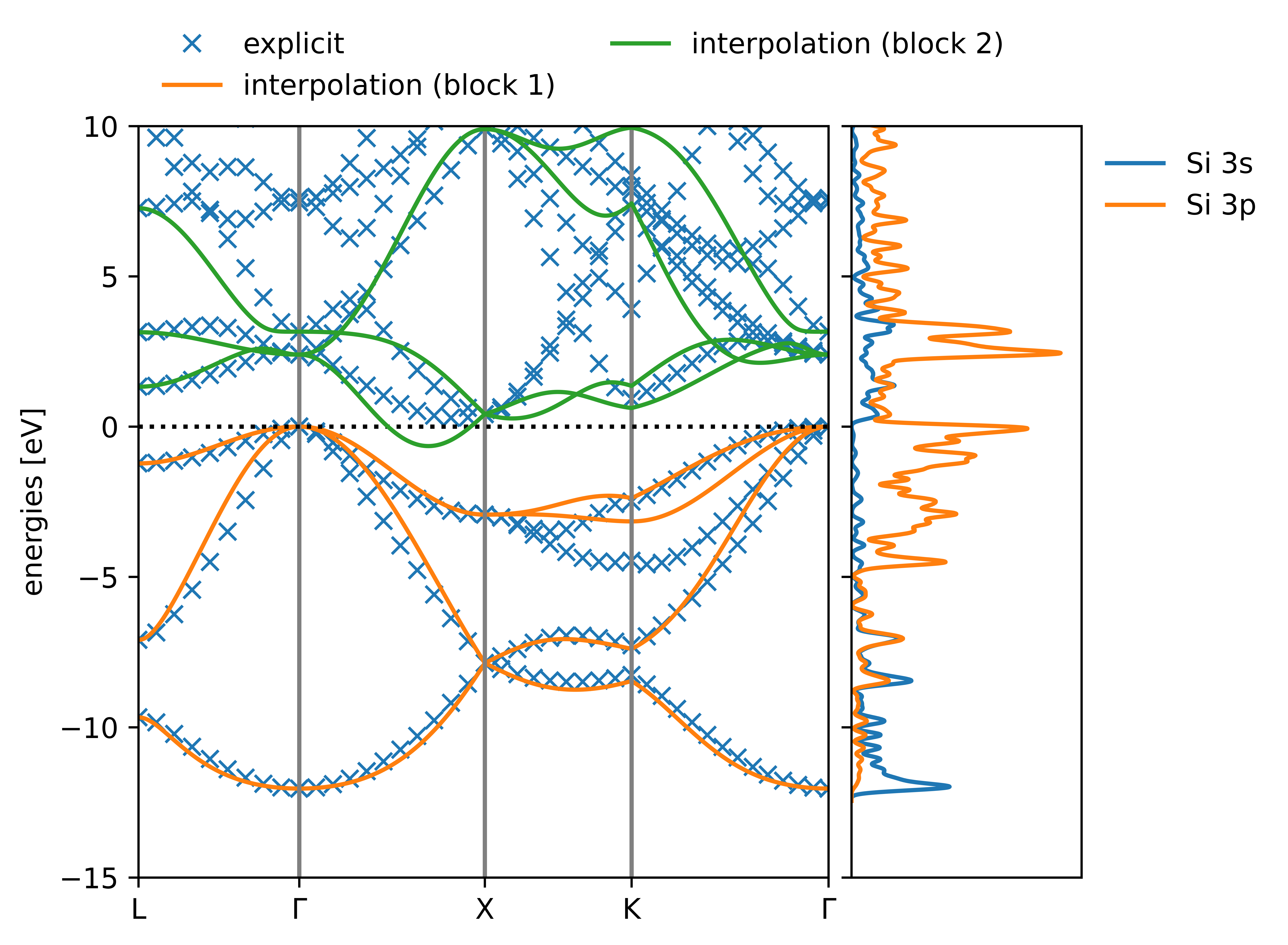
Tutorial 2: the band structure of bulk silicon (calculated via a supercell) — koopmans v1.0.0-beta.6 documentation

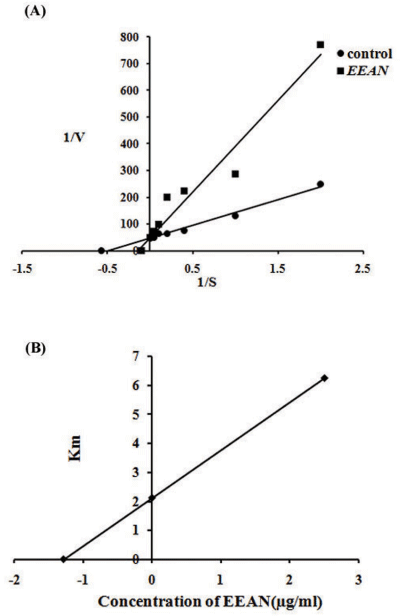
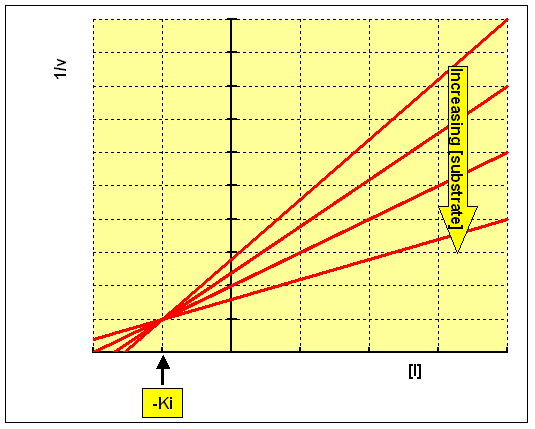
Lineweaver-Burk secondary plot for Ki calculation activity. Compound 3b... | Download Scientific Diagram
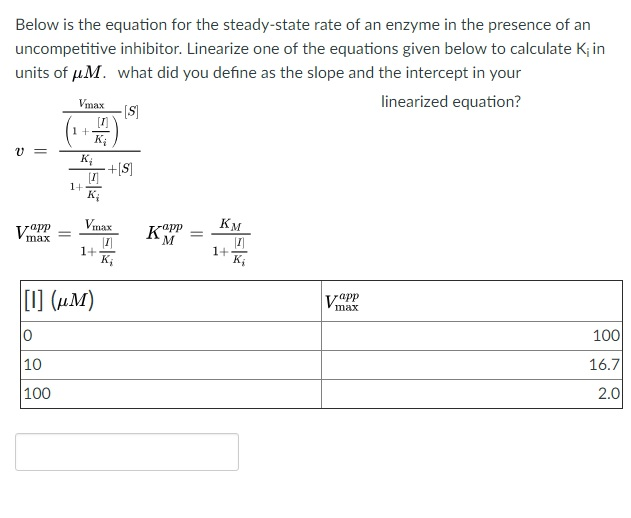
![SOLVED: 17. Calculate Ki for the inhibitor used in the above graph. [I] = 6 µM Ki = µM Please show work. I am not sure how to solve this question. Use SOLVED: 17. Calculate Ki for the inhibitor used in the above graph. [I] = 6 µM Ki = µM Please show work. I am not sure how to solve this question. Use](https://cdn.numerade.com/ask_images/89e65c050ed948c6b17698c7a368b29b.jpg)
SOLVED: 17. Calculate Ki for the inhibitor used in the above graph. [I] = 6 µM Ki = µM Please show work. I am not sure how to solve this question. Use

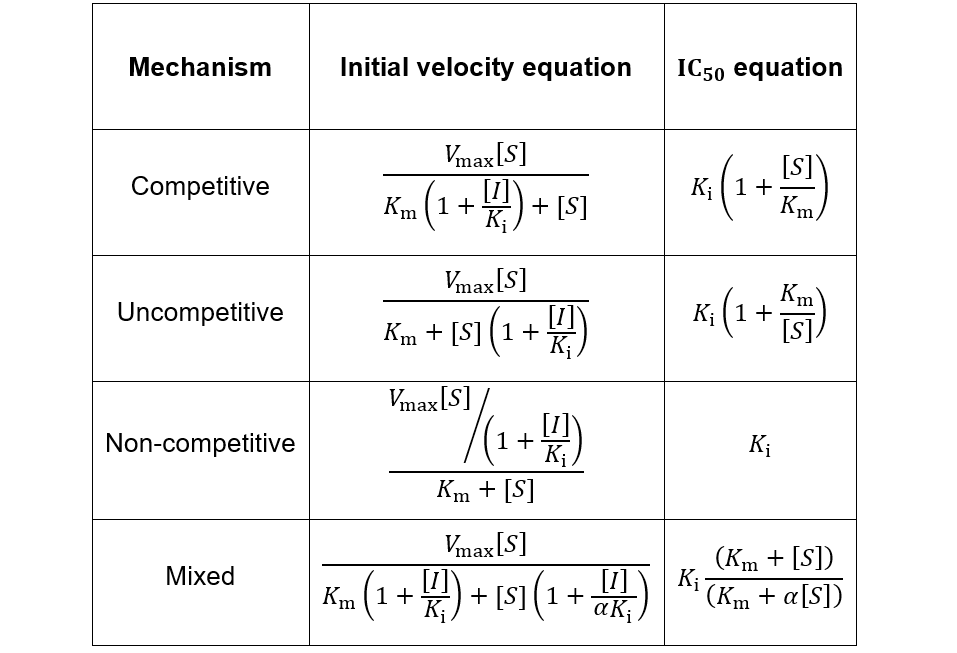
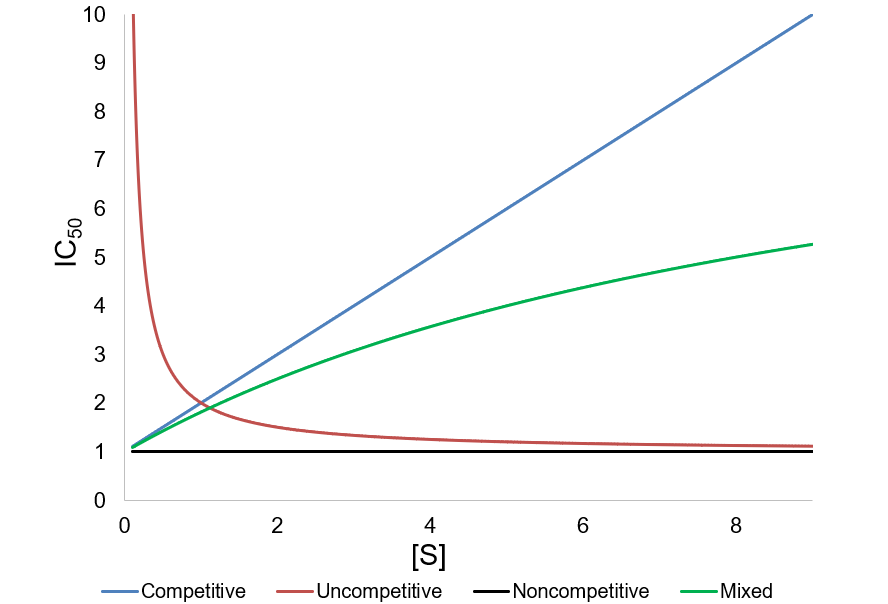
A METHOD FOR IDENTIFICATION OF INHIBITION MECHANISM AND ESTIMATION OF KI IN IN VITRO ENZYME INHIBITION STUDY | Drug Metabolism & Disposition